The secondary analysis of health data is challenging due to confounding, bias, uncertainty, and missingness.
Health data is irregularly sampled in time
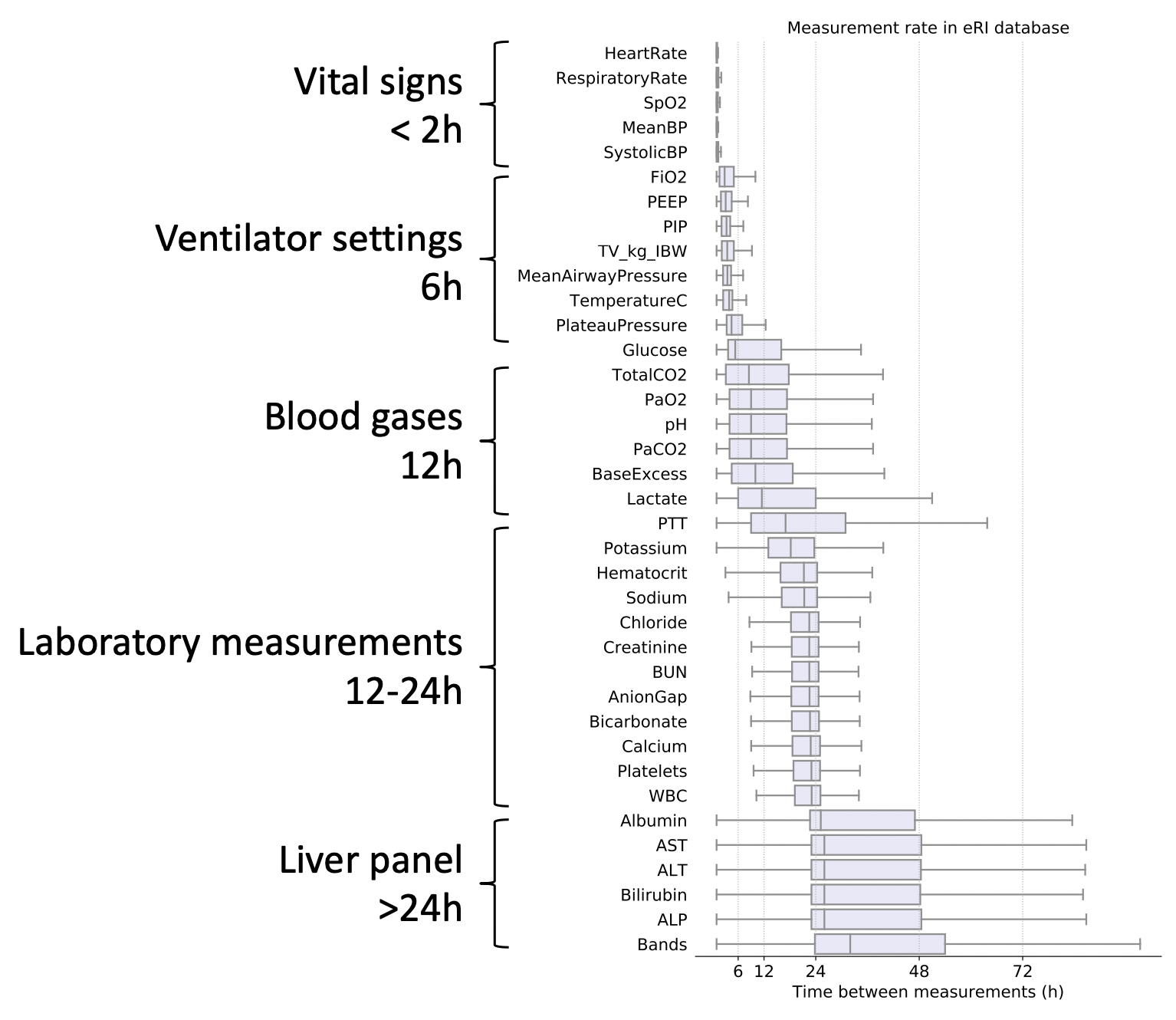
Some data types are more frequently acquired than others. Vitals and laboratory measurmeents are taken for most in-patients. Clinical notes are also usually available for most patients. Ventilator settings, ECG signals, invasive blood pressure measurements, imaging data, and genomics data are are more infrequent. In the intensive care unit (ICU), for instance, continuous signals from ECG and ABP are typically sampled at 125Hz to 500Hz. The raw ECG, ABP, PPG signal can be downsampled to "high-frequency numerics" at 1sec or 1min interval. Some EHR databases will further apply a median filter so the highest sampling rate of vital signs like heart rate, blood pressure, temperature, and oxygen saturation is 5min. Ventilation settings and arterial blood gases can be charted every 6-12hours. Other metabolic and liver panels are often ordered every 12-24hours. These signals are acquired asynchronously, at irregular time intervals, and at different sampling rates.
Adherence to a standard nomenclature
EHR databases may not follow a standard nomenclature (like SNOMED, LOINC, MDIL, etc...), which introduces uncertainty in mapping measurements to standard concepts. OMOP is an example of an EHR database schema that attempts to heavily standardize concepts. However, the publicly accessible MIMIC and Philips eICU databases are not standardized and it is left up to the user to create concept mappings for medications, vital signs, laboratory measurements, and disease diagnoses. Many numeric fields like a laboratory measurement for creatinine can be entered as free text in the EHR user interface, which introduces further noise for the secondary analysis of EHR data.
Critical care data encodes physiology, clinical practice patterns, and clinicians concern
Electronic health records encode more than a patients physiology. The pattern of measurements, lab orders, and treatments capture the clinical decisions made at the bedside. The pattern of clinical decisions changes between institutions, between units in a hospital, the size of the institution (e.g. teaching vs community hospital). Disentangling physiology from clinical concern and care patterns is important to produce generalizable disease prediction models. However, this is not as important when using health data for operational research and to optimize hospital operations (e.g. forecasting bed occupancy).
Mapping patients across care settings
Connecting in-patient data collected in the hospital with out-patient data collected at home or in ambulatory care and emergency medical services is especially challenging. Even critical care data collected in the ICU can have periods of missing data where sensors like ECG, invasive ABP using an arterial line catheter, or PPG are missing for extended periods.
Data reliability
Nurse charted measurements can sometimes be unreliable. As an example, nurse charted respiratory rate at the bedside in the general ward is often rounded to a multiple of 5. Even continuously acquired signals can be unreliable. Timestamps between multiple sensors from different vendors (and sometimes the same vendor) can become desynchronized. The internal clock can drift. Some wearable devices may only save downsampled data instead of the raw signal.
Dataset and concept shift
Data drifts over time, institutions, hospital units, and countries. For example, ventilation settings and disease progression will change as more care givers adopt lung protective ventilation protocols; more patients have permissive hypertension and are on vasopressors in the neuro-ICU than in other units. Models that rely on interventions or physiological variables subject to dataset shift are susceptible to silent failure. Appropriate metrics to track dataset shift, model performance, and a pipeline to retrain and redeploy models are necessary.
Generalizability of machine learning models
Algorithms developed for the general ward (GW) setting may not translate to higher acuity settings, and vice versa. Most patients in the GW are not on continuous monitoring and instead vitals are aperiodically nurse charted. Differences in the severity of illness and treatment patterns are also significantly different. Models should be carefully deployed in settings where they were designed and tested.
Reproducibility
Reproducibility of the data extraction pipeline is important for documentation and verification. This becomes challenging for a large team working on different parts of the data extraction pipeline with tasks like concept mapping, data labeling, cohort selection, and data pre-processing divided among scientists. Start by coordinating on data storage, naming conventions (for code and datasets), and tooling. I have found that documenting the pipeline in single function call helps with posterity and reproducibility.
Actionability
Algorithms that are actionable are tied to a protocol or clinical decision support system that is integrated into the clinical workflow. For example, predicting the that a patient is at high risk of hemodynamic shock on it's own is not useful unless it is tied to a protocol that triggers an alert and a clinical decision support system that recommends appropriate fluids or pressors.